Gene regulatory network inference in long-lived C. elegans reveals modular properties that are predictive of novel ageing genes
Times cited: 6
Suriyalaksh, M, Raimondi, C, Mains, A, Segonds-Pichon, A, Mukhtar, S, Murdoch, S, Aldunate, R, Krueger, F, Guimerà, R, Andrews, S, M. Sales-Pardo, M, Casanueva, O.
iScience
25 (1)
,
103663
(2022).
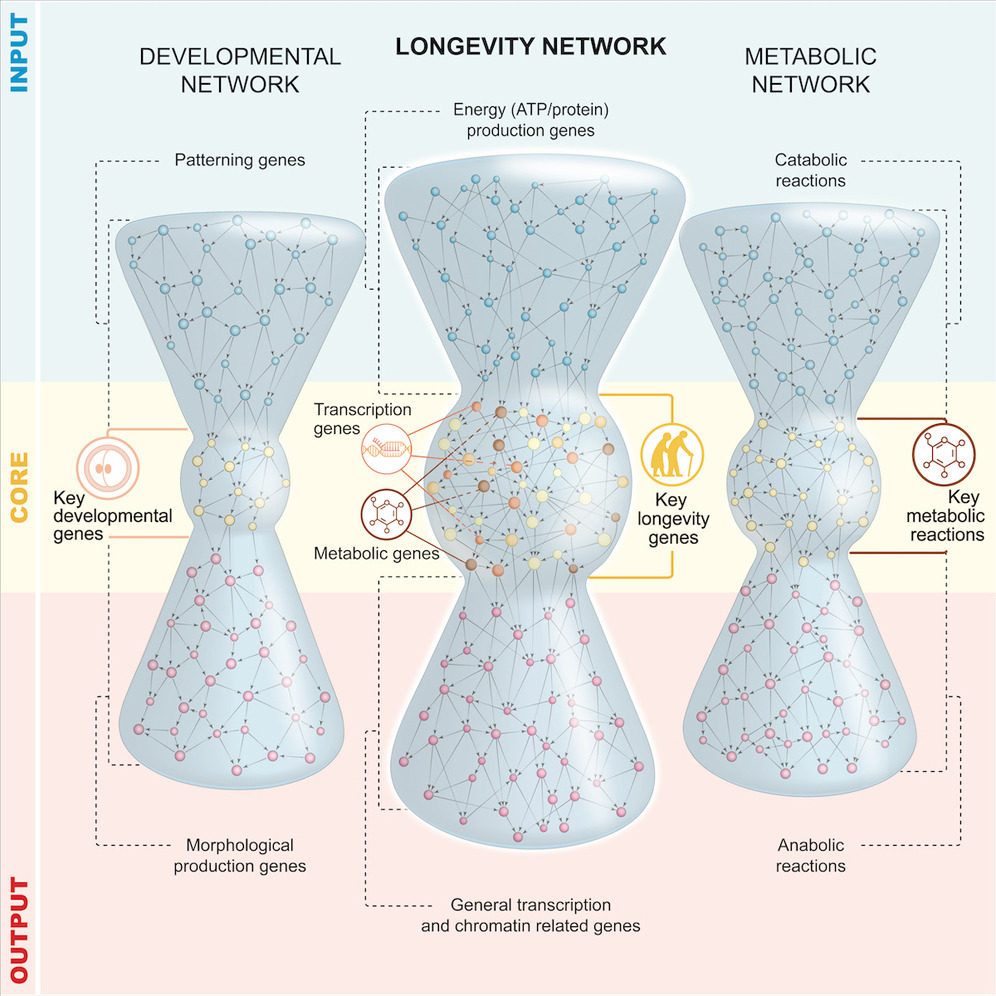
We design a “wisdom-of-the-crowds” GRN inference pipeline, and couple it to complex network analysis, to understand the organisational principles governing gene regulation in long-lived glp-1/Notch C. elegans. The GRN has three layers (input, core, output) and is topologically equivalent to bow-tie/hourglass structures prevalent among metabolic networks. To assess the functional importance of structural layers, we screened 80% of regulators and discovered 50 new ageing genes, 86% with human orthologues. Genes essential for longevity—including ones involved in insulin-like signalling (ILS)—are at the core, indicating that GRN’s structure is predictive of functionality. We used in vivo reporters, and a novel functional network covering 5,497 genetic interactions to make mechanistic predictions. We used genetic epistasis to test some of these predictions, uncovering a novel transcriptional regulator, sup-37 that works alongside DAF-16/FOXO. We present a framework with predictive power that can accelerate discovery in C. elegans and potentially humans.
Media coverage