iMet: A network-based computational tool to assist in the annotation of metabolites from tandem mass spectra
Times cited: 70
Aguilar-Mogas, A, Sales-Pardo, M, Navarro, M, Guimerà, R, Yanes, O.
Anal. Chem.
89 (6)
,
3474
-3482
(2017).
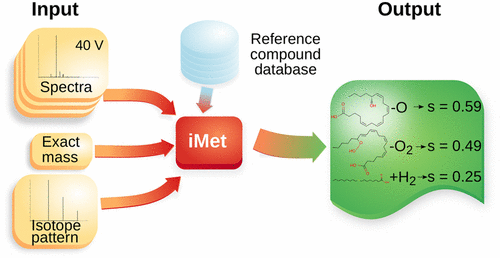
Structural annotation of metabolites relies mainly on tandem mass spectrometry (MS/MS) analysis. However, approximately 90% of the known metabolites reported in metabolomic databases do not have annotated spectral data from standards. This situation has fostered the development of computational tools that predict fragmentation patterns in silico and compare these to experimental MS/MS spectra. However, because such methods require the molecular structure of the detected compound to be available for the algorithm, the identification of novel metabolites in organisms relevant for biotechnological and medical applications remains a challenge. Here, we present iMet, a computational tool that facilitates structural annotation of metabolites not described in databases. iMet uses MS/MS spectra and the exact mass of an unknown metabolite to identify metabolites in a reference database that are structurally similar to the unknown metabolite. The algorithm also suggests the chemical transformation that converts the known metabolites into the unknown one. As a proxy for the structural annotation of novel metabolites, we tested 148 metabolites following a leave-one-out cross-validation procedure or by using MS/MS spectra experimentally obtained in our laboratory. We show that for 89% of the 148 metabolites at least one of the top four matches identified by iMet enables the proper annotation of the unknown metabolites. To further validate iMet, we tested 31 metabolites proposed in the 2012–16 CASMI challenges.